The GruLab group
Led by Sergei Grudinin, CNRS Researcher at Jean Kunzmann Laboratory, Grenoble Alpes University.
- Checkout out GitLab repositories!
Group
Group and collaborators

Sergei Grudinin
CNRS researcher (Research Director), HDR, group leader.
Florian Echelard
PhD student. Development of novel methods in structural bioinformatics for peripheral proteins. Co-supervised with Nathalie Reuter. Funded by the Research Council of Norway.
Dmitrii Zhemchuzhnikov
PhD student. Volumetric data processing. Funded by the Ministry grant.
Khan-Chi Nguyen-Pham
PhD student. Development of language models for virtual drug screening. Co-supervised with Yung-Sing Wong. Funded by Département de Pharmacochimie Moléculaire.
Elodie Laine
Professor at Sorbonne University, Paris, France. Protein sequence to structures and functions. Co-advises V. Lombard and Julien Nguyen Van.
Valentin Lombard
PhD student. Geometric deep manifold learning combined with Natural Language Processing for protein movies. Co-supervised with Elodie Laine. SCAI doctoral grant.
Julien Nguyen Van
PhD student. Deciphering the complexity of proteoform interactions with evolutionary- and physically-informed protein language models. Co-supervised with Elodie Laine. Funded by the ERC.
Emmanuel Jehanno
PhD student. Development of deep-learning methods for material science. Co-supervised with Julien Mairal. Funded by the ERC.
Roman Klypa
PhD student. Novel Generative Models with Equivariant Properties for 3D Biological Data. Co-supervised with Alberto Bietti. Funded by École Polytechnique.
Rémi Vuillemot
PostDoc. Novel methods for integrative structural bioinformatics. Funded by Grenoble University.
Kliment Olechnovič
PostDoc. Development of novel tessellation algorithms. Funded by Marie Skłodowska-Curie Actions.
Davy Darankoum
PhD student. Development of deep-learning methods for EEG signals. Co-supervised with Julien Volle. Funded by SynapCell.Pablo Chacon
Research Staff Scientist at Institute of Physical Chemistry (IQFR-CSIC), Group leader (IQF Madrid, Spain).Nathalie Reuter
Professor at Bergen University (Norway), Group leader.Mikael Lund
Professor at Lund University (Sweden), Group leader.Eric Deeds
Professor at UCLA (USA), Group leader.Julien Mairal
Researcher at Inria, Group leader.Yung-Sing Wong
CNRS Researcher, Group leader.Dina Schneidman
Professor at Hebrew University of Jerusalem, Group leader.We are hiring!
We are looking for highly talented and motivated PhDs and Post-doctoral fellows, with backgrounds in applied mathematics and physics, algorithms and computer science, and structural bioinformatics.
Get in touch!Software
Try them out
- All
- Scattering
- Symmetries
- Motions
- Proteins
- [R,D]NAs
- Drugs
- Design
- Algorithms
- ML
- DL
NOLB Normal Modes[Video]
How does my favourite protein move?
S. Grudinin+, E. Laine+, and A. Hoffmann (2020) Biophys. J. A. Hoffmann and S. Grudinin (2017) J. Chem. Th. Comput.Publications
Full list here
| Title | Year | |
|---|---|---|
|
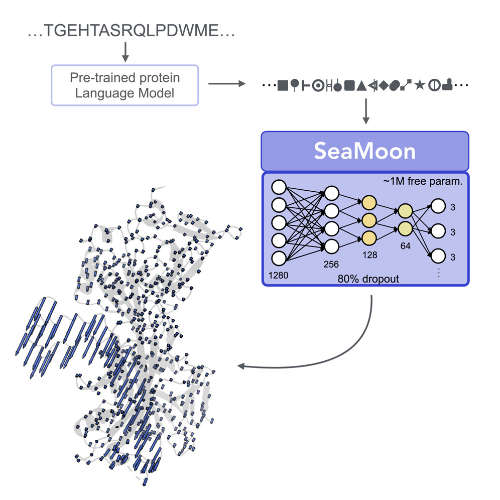
SeaMoon: From protein language models to continuous structural heterogeneity
V Lombard, D Timsit, S Grudinin, E Laine
Structure, 2025
|
2025 | |
|
Voronota‐LT: Efficient, Flexible, and Solvent‐Aware Tessellation‐Based Analysis of Atomic Interactions K Olechnovič, S Grudinin Journal of Computational Chemistry 46 (19), e70178, 2025 | 2025 | |
| Atomic Conformational Dynamics and Actin-Crosslinking Function of Alpha-Actinin Revealed by SimHS-AFMfit KX Ngo, T Sumikama, R Vuillemot, HG Nguyen, TQP Uyeda, NTP Le, ... bioRxiv, 2025.04. 06.647477, 2025 | 2025 | |
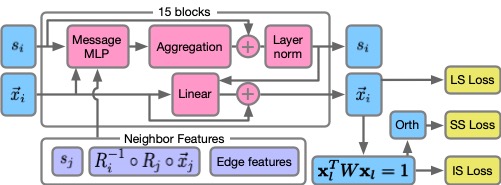
| PETIMOT: A Novel Framework for Inferring Protein Motions from Sparse Data Using SE (3)-Equivariant Graph Neural Networks V Lombard, S Grudinin, E Laine arXiv preprint arXiv:2504.02839, 2025 | 2025 | |
| DynamicGT: a dynamic-aware geometric transformer model to predict protein binding interfaces in flexible and disordered regions O Mokhtari, S Grudinin, Y Karami, H Khakzad bioRxiv, 2025.03. 04.641377, 2025 | 1 | 2025 |
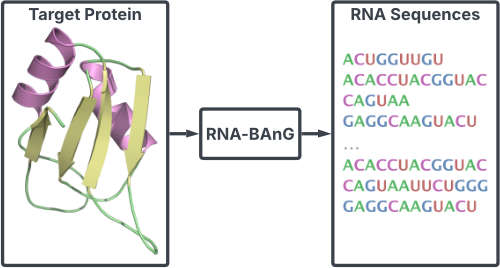
| BAnG: Bidirectional Anchored Generation for Conditional RNA Design R Klypa, A Bietti, S Grudinin arXiv preprint arXiv:2502.21274, 2025 | 2025 | |
| Application of Protein Language Models to Drug Design NP Khanh-Chi, YS Wong, S Grudinin Research Day 2024-Groupe de Travail LEGO-Machine Learning for genomics, 2024 | 2024 | |
| From epilepsy seizures classification to detection: A deep learning-based approach for raw EEG signals D Darankoum, M Villalba, C Allioux, B Caraballo, C Dumont, E Gronlier, ... arXiv preprint arXiv:2410.03385, 2024 | 6 | 2024 |
| SeaMoon: Prediction of molecular motions based on language models V Lombard, D Timsit, S Grudinin, E Laine bioRxiv, 2024.09. 23.614585, 2024 | 2024 | |
| Anisotropic coarse-grain Monte Carlo simulations of lysozyme, lactoferrin, and NISTmAb by precomputing atomistic models HW Hatch, C Bergonzo, MA Blanco, G Yuan, S Grudinin, M Lund, ... The Journal of Chemical Physics 161 (9), 2024 | 6 | 2024 |
| Assessment of four theoretical approaches to predict protein flexibility in the crystal phase and solution ŁJ Dziadek, AK Sieradzan, C Czaplewski, M Zalewski, F Banaś, M Toczek, ... Journal of Chemical Theory and Computation 20 (17), 7667-7681, 2024 | 1 | 2024 |
| ILPO-NET: Network for the invariant recognition of arbitrary volumetric patterns in 3D D Zhemchuzhnikov, S Grudinin Joint European Conference on Machine Learning and Knowledge Discovery in …, 2024 | 2 | 2024 |
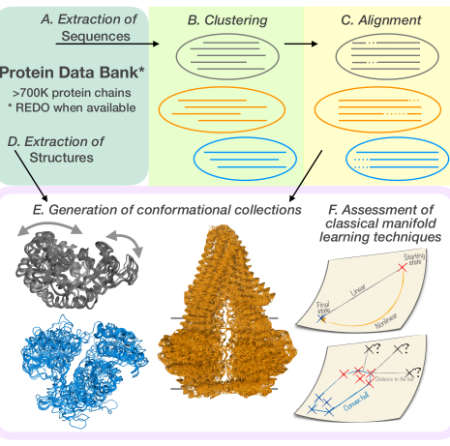
| Explaining conformational diversity in protein families through molecular motions V Lombard, S Grudinin, E Laine Scientific Data 11 (1), 752, 2024 | 11 | 2024 |
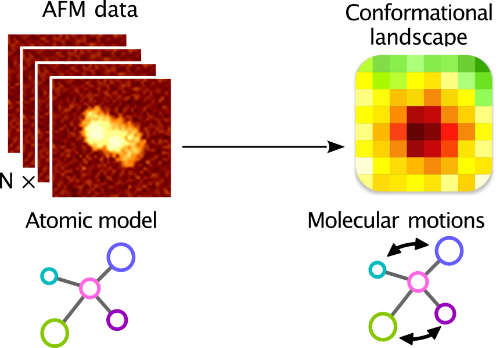
| AFMfit: Deciphering conformational dynamics in AFM data using fast nonlinear NMA and FFT-based search R Vuillemot, JL Pellequer, S Grudinin bioRxiv, 2024.06. 03.597083, 2024 | 4 | 2024 |
| On the Fourier analysis in the SO (3) space: EquiLoPO Network D Zhemchuzhnikov, S Grudinin arXiv preprint arXiv:2404.15979, 2024 | 1 | 2024 |
| New Challenges in Structural Bioinformatics: When Physics Meets Big Data S Grudinin Université Grenoble Alpes, 2024 | 2024 | |
| Ocena czterech teoretycznych podejść do przewidywania elastyczności białek w fazie krystalicznej iw roztworze Ł Dziadek, A Sieradzan, C Czaplewski, M Zalewski, F Banaś, M Toczek, ... Doctoral School of Natural Sciences, 2024 | 2024 | |
| Annotating Macromolecular Complexes in the Protein Data Bank: Improving the FAIRness of Structure Data SD Appasamy, J Berrisford, R Gaborova, S Nair, S Anyango, S Grudinin, ... Scientific data 10 (1), 853, 2023 | 3 | 2023 |
| Stages of OCP–FRP Interactions in the Regulation of Photoprotection in Cyanobacteria, Part 2: Small-Angle Neutron Scattering with Partial Deuteration M Golub, M Moldenhauer, O Matsarskaia, A Martel, S Grudinin, ... The Journal of Physical Chemistry B 127 (9), 1901-1913, 2023 | 8 | 2023 |
| Docking-based long timescale simulation of cell-size protein systems at atomic resolution IA Vakser, S Grudinin, NW Jenkins, PJ Kundrotas, EJ Deeds Proceedings of the National Academy of Sciences 119 (41), e2210249119, 2022 | 16 | 2022 |
| 6DCNN with roto-translational convolution filters for volumetric data processing D Zhemchuzhnikov, I Igashov, S Grudinin Proceedings of the AAAI Conference on Artificial Intelligence 36 (4), 4707-4715, 2022 | 4 | 2022 |
| 3D live cell imaging of whole organoids in time-lapse using intensity diffraction tomography W Pierré, L Hervé, C Allier, S Morales, S Grudinin, P RAY, C Arnoult, ... Unconventional Optical Imaging III, PC1213612, 2022 | 2022 | |
| 3D time-lapse imaging of a mouse embryo using intensity diffraction tomography embedded inside a deep learning framework W Pierré, L Hervé, C Paviolo, O Mandula, V Remondiere, S Morales, ... Applied optics 61 (12), 3337-3348, 2022 | 20 | 2022 |
| Modeling SARS‐CoV‐2 proteins in the CASP‐commons experiment A Kryshtafovych, J Moult, WM Billings, D Della Corte, K Fidelis, S Kwon, ... Proteins: Structure, Function, and Bioinformatics 89 (12), 1987-1996, 2021 | 22 | 2021 |
| Prediction of protein assemblies, the next frontier: The CASP14‐CAPRI experiment MF Lensink, G Brysbaert, T Mauri, N Nadzirin, S Velankar, RAG Chaleil, ... Proteins: Structure, Function, and Bioinformatics 89 (12), 1800-1823, 2021 | 121 | 2021 |
| Protein sequence‐to‐structure learning: Is this the end (‐to‐end revolution)? E Laine, S Eismann, A Elofsson, S Grudinin Proteins: Structure, Function, and Bioinformatics 89 (12), 1770-1786, 2021 | 43 | 2021 |
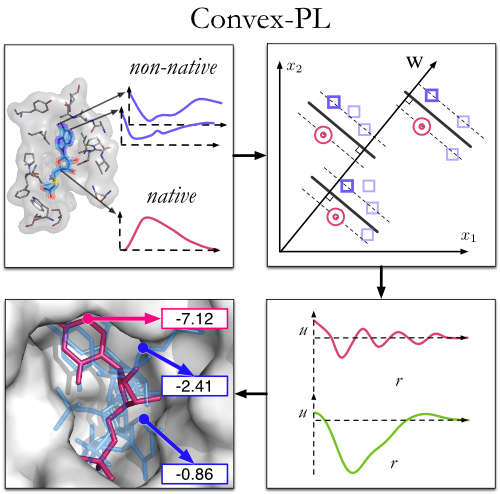
| Convex-PLR – Revisiting affinity predictions and virtual screening using physics-informed machine learning M Kadukova, V Chupin, S Grudinin bioRxiv, 2021.09. 13.460049, 2021 | 1 | 2021 |
| VoroCNN: deep convolutional neural network built on 3D Voronoi tessellation of protein structures I Igashov, K Olechnovič, M Kadukova, Č Venclovas, S Grudinin Bioinformatics 37 (16), 2332-2339, 2021 | 38 | 2021 |
| Deep learning entering the post-protein structure prediction era: new horizons for structural biology S Grudinin Acta Crystallographica Section A: Foundations and Advances [2014-...] 77 (C473), 2021 | 2021 | |
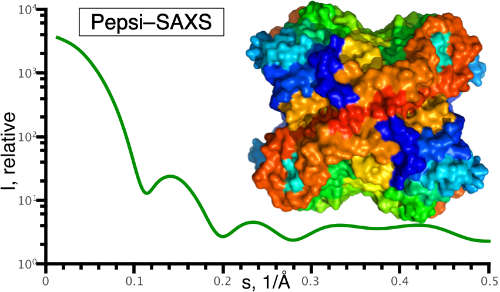
| Pepsi-SAXS/SANS–small-angle scattering guided tools for integrative structural bioinformatics S Grudinin, A Martel, S Prevost Acta Crystallogr. A Found. Adv. A 77, C49, 2021 | 3 | 2021 |
| Spherical convolutions on molecular graphs for protein model quality assessment I Igashov, N Pavlichenko, S Grudinin Machine Learning: Science and Technology 2 (4), 045005, 2021 | 22 | 2021 |
| KORP-PL: a coarse-grained knowledge-based scoring function for protein–ligand interactions M Kadukova, KS Machado, P Chacón, S Grudinin Bioinformatics 37 (7), 943-950, 2021 | 31 | 2021 |
| HOPMA: Boosting protein functional dynamics with colored contact maps E Laine, S Grudinin The Journal of Physical Chemistry B 125 (10), 2577-2588, 2021 | 12 | 2021 |
| Deep learning framework applied to optical diffraction tomography (ODT) W Pierré, L Hervé, C Allier, S Morales, S Grudinin, S Chowdhury, L Waller, ... Three-Dimensional and Multidimensional Microscopy: Image Acquisition and …, 2021 | 2 | 2021 |
| Session introduction: AI-driven Advances in Modeling of Protein Structure K Fidelis, S Grudinin Pacific Symposium on Biocomputing 2022, 1-9, 2021 | 2021 | |
| Hydroxylation of antitubercular drug candidate, SQ109, by mycobacterial cytochrome P450 S Bukhdruker, T Varaksa, I Grabovec, E Marin, P Shabunya, M Kadukova, ... International journal of molecular sciences 21 (20), 7683, 2020 | 20 | 2020 |
| KORP-PL: a coarse-grained knowledge-based scoring function for protein-ligand interactions P Chacón, M Kadukova, K Santos Machado, S Grudinin Oxford University Press, 2020 | 2020 | |
| Interdomain flexibility within NADPH oxidase suggested by SANS using LMNG stealth carrier A Vermot, I Petit-Härtlein, C Breyton, A Le Roy, M Thépaut, C Vivès, ... Biophysical journal 119 (3), 605-618, 2020 | 11 | 2020 |
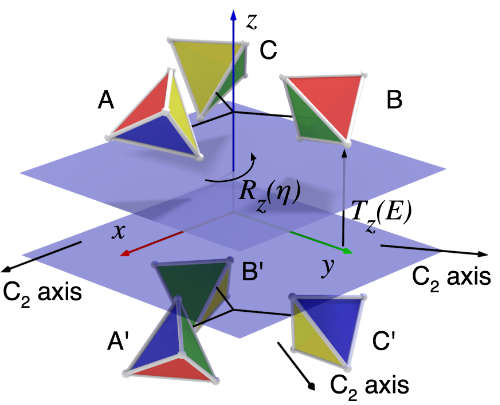
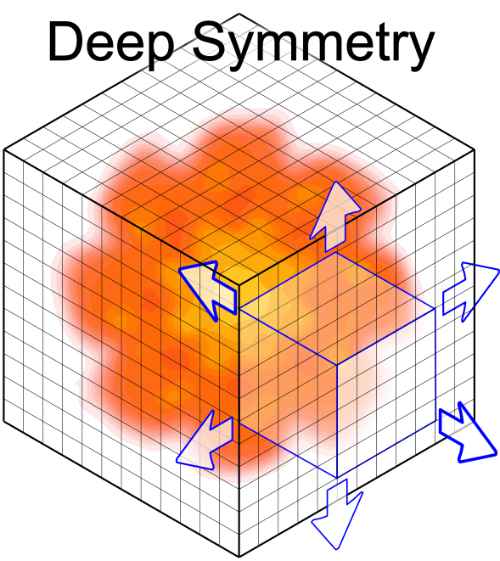
| AnAnaS: software for analytical analysis of symmetries in protein structures G Pagès, S Grudinin Protein Structure Prediction, 245-257, 2020 | 10 | 2020 |
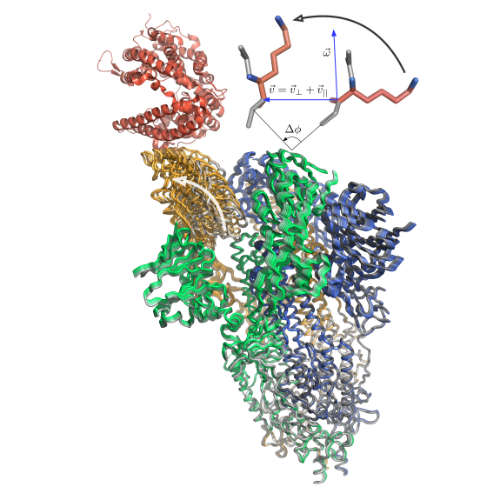
| Predicting protein functional motions: an old recipe with a new twist S Grudinin, E Laine, A Hoffmann Biophysical journal 118 (10), 2513-2525, 2020 | 17 | 2020 |
| Combining molecular dynamics simulations with small-angle X-ray and neutron scattering data to study multi-domain proteins in solution AH Larsen, Y Wang, S Bottaro, S Grudinin, L Arleth, K Lindorff-Larsen PLoS computational biology 16 (4), e1007870, 2020 | 108 | 2020 |
| 16–17 Mar 2020 ILL4 Europe/Paris timezone A Karczynska Europe 16, 17, 2020 | 2020 | |
| Docking rigid macrocycles using Convex-PL, AutoDock Vina, and RDKit in the D3R Grand Challenge 4 M Kadukova, V Chupin, S Grudinin Journal of Computer-Aided Molecular Design 34 (2), 191-200, 2020 | 14 | 2020 |
| Blind prediction of homo‐and hetero‐protein complexes: The CASP13‐CAPRI experiment MF Lensink, G Brysbaert, N Nadzirin, S Velankar, RAG Chaleil, T Gerguri, ... Proteins: Structure, Function, and Bioinformatics 87 (12), 1200-1221, 2019 | 137 | 2019 |
| Small angle X‐ray scattering‐assisted protein structure prediction in CASP13 and emergence of solution structure differences GL Hura, CD Hodge, D Rosenberg, D Guzenko, JM Duarte, ... Proteins: Structure, Function, and Bioinformatics 87 (12), 1298-1314, 2019 | 33 | 2019 |
| Assessment of chemical‐crosslink‐assisted protein structure modeling in CASP13 JE Fajardo, R Shrestha, N Gil, A Belsom, SN Crivelli, C Czaplewski, ... Proteins: Structure, Function, and Bioinformatics 87 (12), 1283-1297, 2019 | 33 | 2019 |
| Controlled‐advancement rigid‐body optimization of nanosystems P Popov, S Grudinin, A Kurdiuk, P Buslaev, S Redon Journal of computational chemistry 40 (27), 2391-2399, 2019 | 1 | 2019 |
| Smooth orientation-dependent scoring function for coarse-grained protein quality assessment M Karasikov, G Pagès, S Grudinin Bioinformatics 35 (16), 2801-2808, 2019 | 70 | 2019 |
| KAP1 is an antiparallel dimer with a functional asymmetry G Fonti, MJ Marcaida, LC Bryan, S Träger, AS Kalantzi, PYJL Helleboid, ... Life science alliance 2 (4), 2019 | 31 | 2019 |
| Symmetry in protein complexes S Grudinin, G Pagès, DW Ritchie F1000Research 8, 2019 | 2019 | |
| DeepSymmetry: using 3D convolutional networks for identification of tandem repeats and internal symmetries in protein structures G Pagès, S Grudinin Bioinformatics, btz454, 2019 | 14 | 2019 |
| KAP1 is an antiparallel dimer with a functional asymmetry DD Helleboid, M Tully, S Grudinin, D Trono, B Fierz, M Dal | 2019 | |
| Protein model quality assessment using 3D oriented convolutional neural networks G Pagès, B Charmettant, S Grudinin Bioinformatics, 2019 | 112 | 2019 |
| MS12-P03| SYMMETRY IN STRUCTURES OF PROTEIN ASSEMBLIES S Grudinin, G Pages Acta Cryst 75, e187, 2019 | 2019 | |
| Knowledge-based prediction of protein-ligand binding affinities M Kadukova, V Chupin, S Grudinin JOURNAL OF BIOENERGETICS AND BIOMEMBRANES 50 (6), 546-547, 2018 | 2018 | |
| Deep convolutional networks for quality assessment of protein folds G Derevyanko, S Grudinin, Y Bengio, G Lamoureux Bioinformatics 34 (23), 4046-4053, 2018 | 134 | 2018 |
| Analytical symmetry detection in protein assemblies. II. Dihedral and cubic symmetries G Pagès, S Grudinin Journal of structural biology 203 (3), 185-194, 2018 | 22 | 2018 |
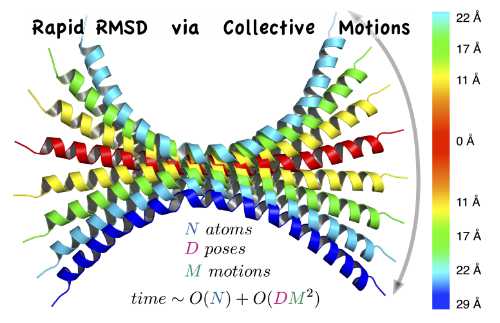
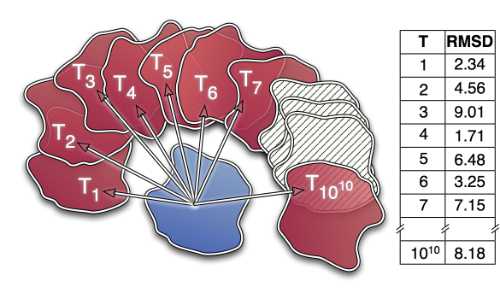
| RapidRMSD: rapid determination of RMSDs corresponding to motions of flexible molecules E Neveu, P Popov, A Hoffmann, A Migliosi, X Besseron, G Danoy, ... Bioinformatics 34 (16), 2757-2765, 2018 | 29 | 2018 |
| BioSAXS at ESRF: The solution scattering Beam line BM29 S Grudinin, A Hoffmann, A Martel, S Prevost ACTA CRYSTALLOGRAPHICA A-FOUNDATION AND ADVANCES 74, E179-E179, 2018 | 2018 | |
| Analytical symmetry detection in protein assemblies. I. Cyclic symmetries G Pagès, E Kinzina, S Grudinin Journal of Structural Biology 203 (2), 142-148, 2018 | 24 | 2018 |
| Eurecon: equidistant uniform rigid-body ensemble constructor P Popov, S Grudinin Journal of Molecular Graphics and Modelling 80, 313-319, 2018 | 5 | 2018 |
| User Guide for NOLB: Non-Linear Normal Mode Analysis S Grudinin | 2018 | |
| CryoEM models and associated data submitted to the 2015/2016 EMDataBank model challenge CL Lawson, A Kryshtafovych, W Chiu, P Adams, A Brünger, G Kleywegt, ... (No Title), 2018 | 6 | 2018 |
| Protein model quality assessment using 3D oriented convolutional neural networks. bioRxiv G Pages, B Charmettant, S Grudinin | 2 | 2018 |
| Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2 M Kadukova, S Grudinin Journal of computer-aided molecular design 32 (1), 151-162, 2018 | 23 | 2018 |
| Deep convolutional networks for protein structure quality assessment G Derevyanko, G Lamoureux, S Grudinin PROTEIN SCIENCE 26, 111-111, 2017 | 2017 | |
| User Guide for AnAnaS: Analytical Analyzer of Symmetries G Pagès, S Grudinin | 2017 | |
| Interaction parameters for the input set of molecular structures G Cheremovsky, P Popov, G Derevyanko, S Grudinin US Patent App. 15/529,774, 2017 | 4 | 2017 |
| Convex-PL: a novel knowledge-based potential for protein-ligand interactions deduced from structural databases using convex optimization M Kadukova, S Grudinin Journal of computer-aided molecular design 31 (10), 943-958, 2017 | 46 | 2017 |
| A novel fast Fourier transform accelerated off-grid exhaustive search method for cryo-electron microscopy fitting A Hoffmann, V Perrier, S Grudinin Applied Crystallography 50 (4), 1036-1047, 2017 | 6 | 2017 |
| Mechanism of transmembrane signaling by sensor histidine kinases I Gushchin, I Melnikov, V Polovinkin, A Ishchenko, A Yuzhakova, ... Science 356 (6342), eaah6345, 2017 | 171 | 2017 |
| NOLB: Nonlinear rigid block normal-mode analysis method A Hoffmann, S Grudinin Journal of chemical theory and computation 13 (5), 2123-2134, 2017 | 80 | 2017 |
| Pepsi-SAXS: an adaptive method for rapid and accurate computation of small-angle X-ray scattering profiles S Grudinin, M Garkavenko, A Kazennov Biological Crystallography 73 (5), 449-464, 2017 | 153 | 2017 |
| Using machine learning to predict protein-ligand interactions M Kadukova, S Grudinin Understanding Protein Interactions: from Molecules to Organisms, 16, 2017 | 2017 | |
| Modeling and minimizing CAPRI round 30 symmetrical protein complexes from CASP‐11 structural models M El Houasli, B Maigret, MD Devignes, AW Ghoorah, S Grudinin, ... Proteins: Structure, Function, and Bioinformatics 85 (3), 463-469, 2017 | 3 | 2017 |
| New insights on signal propagation by sensory rhodopsin II/transducer complex A Ishchenko, E Round, V Borshchevskiy, S Grudinin, I Gushchin, JP Klare, ... Scientific reports 7 (1), 41811, 2017 | 32 | 2017 |
| Quadratic Programming Approach to Fit Protein Complexes into Electron Density Maps R Pogodin, A Katrutsa, S Grudinin arXiv preprint arXiv:1701.02192, 2017 | 2017 | |
| Inverse protein folding problem via quadratic programming A Riazanov, M Karasikov, S Grudinin arXiv preprint arXiv:1701.00673, 2017 | 4 | 2017 |
| AnAnaS: Analytical Analyzer of Symmetries G Pagès, S Grudinin | 2017 | |
| Search of new optogenetics tools by means of structural and functional characterization of novel microbial rhodopsins which reproduce mutations of already known ones I Okhrimenko, P Popov, S Pakhomova, V Zyulina, G Legkun, N Malyar, ... FEBS JOURNAL 283, 219-219, 2016 | 1 | 2016 |
| PEPSI-Dock: a detailed data-driven protein–protein interaction potential accelerated by polar Fourier correlation E Neveu, DW Ritchie, P Popov, S Grudinin Bioinformatics 32 (17), i693-i701, 2016 | 27 | 2016 |
| Predicting binding poses and affinities for protein-ligand complexes in the 2015 D3R Grand Challenge using a physical model with a statistical parameter estimation S Grudinin, M Kadukova, A Eisenbarth, S Marillet, F Cazals Journal of computer-aided molecular design 30 (9), 791-804, 2016 | 21 | 2016 |
| Prediction of homoprotein and heteroprotein complexes by protein docking and template‐based modeling: a CASP‐CAPRI experiment MF Lensink, S Velankar, A Kryshtafovych, SY Huang, ... Proteins: Structure, Function, and Bioinformatics 84, 323-348, 2016 | 173 | 2016 |
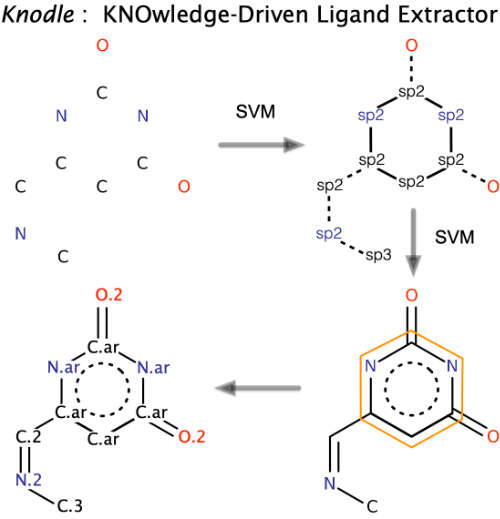
| Knodle: a support vector machines-based automatic perception of organic molecules from 3D coordinates M Kadukova, S Grudinin Journal of Chemical Information and Modeling 56 (8), 1410-1419, 2016 | 24 | 2016 |
| Predicting binding poses and affinities in the CSAR 2013–2014 docking exercises using the knowledge-based Convex-PL potential S Grudinin, P Popov, E Neveu, G Cheremovskiy Journal of Chemical Information and Modeling 56 (6), 1053-1062, 2016 | 15 | 2016 |
| Principal component analysis of lipid molecule conformational changes in molecular dynamics simulations P Buslaev, V Gordeliy, S Grudinin, I Gushchin Journal of chemical theory and computation 12 (3), 1019-1028, 2016 | 47 | 2016 |
| Spherical polar Fourier assembly of protein complexes with arbitrary point group symmetry DW Ritchie, S Grudinin Applied Crystallography 49 (1), 158-167, 2016 | 57 | 2016 |
| KNODLE-A MACHINE LEARNING-BASED TOOL FOR PERCEPTION OF ORGANIC MOLECULES FROM 3D COORDINATES MN Kadukova, S Grudinin ХХ Менделеевский съезд по общей и прикладной химии, 138-138, 2016 | 2016 | |
| Knowledge-based prediction model for characterization of microbial rhodopsins for optogenetics A Ushakov, S Grudinin, I Okhrimenko FEBS J 283 (1), 2-014, 2016 | 3 | 2016 |
| Knowledge of native protein–protein interfaces is sufficient to construct predictive models for the selection of binding candidates P Popov, S Grudinin Journal of chemical information and modeling 55 (10), 2242-2255, 2015 | 24 | 2015 |
| Advances in GPCR modeling evaluated by the GPCR Dock 2013 assessment: meeting new challenges I Kufareva, V Katritch, RC Stevens, R Abagyan Structure 22 (8), 1120-1139, 2014 | 195 | 2014 |
| HermiteFit: fast-fitting atomic structures into a low-resolution density map using three-dimensional orthogonal Hermite functions G Derevyanko, S Grudinin Biological Crystallography 70 (8), 2069-2084, 2014 | 10 | 2014 |
| Unique DC-SIGN clustering activity of a small glycomimetic: A lesson for ligand design I Sutkeviciute, M Thépaut, S Sattin, A Berzi, J McGeagh, S Grudinin, ... ACS chemical biology 9 (6), 1377-1385, 2014 | 62 | 2014 |
| X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism P Nogly, I Gushchin, A Remeeva, AM Esteves, N Borges, P Ma, ... Nature Communications 5 (1), 4169, 2014 | 51 | 2014 |
| Rapid determination of RMSDs corresponding to macromolecular rigid body motions P Popov, S Grudinin Journal of computational chemistry 35 (12), 950-956, 2014 | 28 | 2014 |
| Blind prediction of interfacial water positions in CAPRI MF Lensink, IH Moal, PA Bates, PL Kastritis, ASJ Melquiond, E Karaca, ... Proteins: Structure, Function, and Bioinformatics 82 (4), 620-632, 2014 | 64 | 2014 |
| DockTrina: Docking triangular protein trimers P Popov, DW Ritchie, S Grudinin Proteins: Structure, Function, and Bioinformatics 82 (1), 34-44, 2014 | 23 | 2014 |
| Community‐wide evaluation of methods for predicting the effect of mutations on protein–protein interactions R Moretti, SJ Fleishman, R Agius, M Torchala, PA Bates, PL Kastritis, ... Proteins: Structure, Function, and Bioinformatics 81 (11), 1980-1987, 2013 | 115 | 2013 |
| Two distinct states of the HAMP domain from sensory rhodopsin transducer observed in unbiased molecular dynamics simulations I Gushchin, V Gordeliy, S Grudinin PLoS One 8 (7), e66917, 2013 | 23 | 2013 |
| Ground state structure of D75N mutant of sensory rhodopsin II in complex with its cognate transducer A Ishchenko, E Round, V Borshchevskiy, S Grudinin, I Gushchin, JP Klare, ... Journal of Photochemistry and Photobiology B: Biology 123, 55-58, 2013 | 13 | 2013 |
Conferences
Upcoming Scientific Events
Contact
Contact Us
Location:
LJK, UMR 5224
CNRS - Grenoble Alpes University
Bâtiment IMAG - first floor
150 place du Torrent
38401 Saint Martin d'Hères, FRANCE
Email:
sergei.grudinin@univ-grenoble-alpes.fr